A ‘behind-the research’ conversation with René L. Warren on unravelling the genetic clues to COVID-19 severity
By: Shantala Hari Dass

As the world grappled with the complexities of the COVID-19 pandemic, scientists raced to comprehend the factors influencing the severity of the disease. Intrigued by the spectrum of COVID-19 severity, researchers from Dr. Inanc Birol’s group at Vancouver’s Michael Smith Genome Sciences Centre, led by Bioinformatics Team Leader René L. Warren, embarked on a quest to understand the spectrum of COVID-19 severity, including possible links to immune genes.
The Human Leukocyte Antigens (HLAs) are such genes and, for the central role they play in human immunity, they are disease determinants. Because versions of HLA genes (called HLA alleles) are found at different frequencies within human populations, they could be a molecular clue to explain why certain groups might be more susceptible to infection and exhibit more or less severe symptoms. HLAs can be likened to servers presenting food in a restaurant. When the body encounters a pathogen, it breaks it down into protein fragments, presented by HLA ‘servers’ to immune cells, triggering an immune response. However, these ‘servers’ are selective in what they present. Several HLA variants, such as HLA-C*04:01, bind to viral degradation fragments from the SARS family, which includes the coronavirus responsible for COVID-19. In our analogy of ‘server’ genes, HLA-C*04:01, contrary to most HLA variants, can be seen as serving only a restricted number of viral fragments to the host immune system, potentially limiting the immune system’s ability to combat the COVID-19 virus.
At the start of the COVID-19 pandemic, multiple research groups began probing for possible associations between HLAs and COVID-19 disease outcomes. Such studies had to march in real-time with efforts to collect data and build datasets. Warren aimed to investigate whether our HLA makeup influences susceptibility to and severity of COVID-19, and they had the exact tool to do it.
“We had developed a bioinformatics tool called HLAminer a decade ago, aimed at determining HLA types from next-generation sequencing datasets. We saw potential in applying this tool to explore this research question,” Warren explains
The research team began analyzing existing data, starting with whole genome sequences from eight COVID-19 patients at the Wuhan seafood market, leading to the first publication of this research in 2020. They expanded their analyses to include a genomic dataset with meta/clinical variables from COVID-19 patients in New York. While these initial studies revealed an intriguing (but weak) association between an HLA allele-C*04:01- and severe COVID-19, they were limited by the small sample sizes of 100 patients.Since the initial report of an association between HLA-C*04:01 alleles and COVID-19 severity, many other research teams have confirmed these findings in patient cohorts worldwide, including in Europe, India, Armenia, Spain, and the United Arab Emirates. However, these studies were also limited by small (in the range of 100s) sample sizes.
Since the initial report of an association between HLA-C*04:01 alleles and COVID-19 severity, many other research teams have confirmed these findings in patient cohorts worldwide, including in Europe, India, Armenia, Spain, and the United Arab Emirates. However, these studies were also limited by small (in the range of 100s) sample sizes.
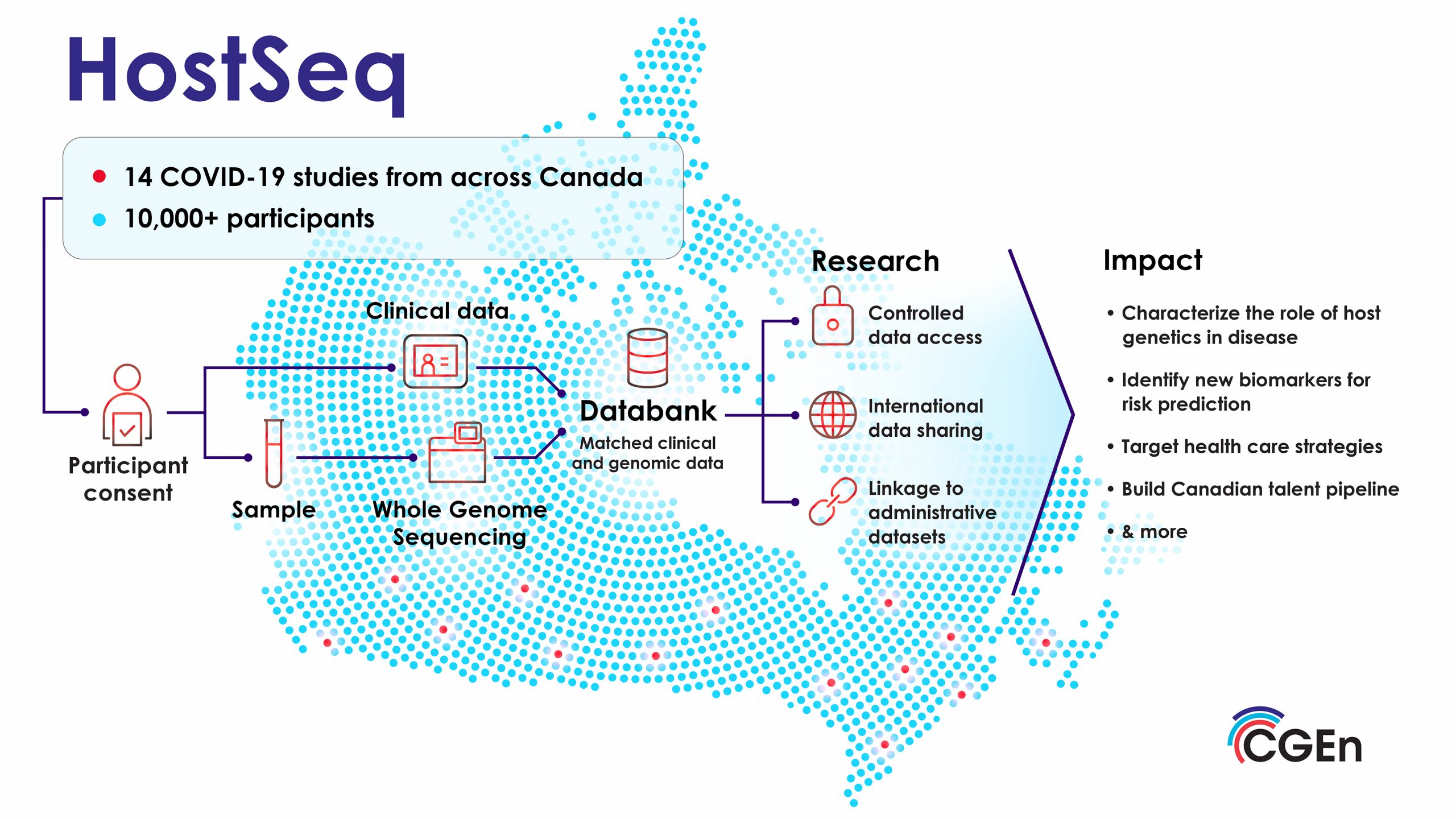
A pivotal step for Warren was analyzing the HostSeq dataset. HostSeq is a first-of-its-kind Canadian genomic and clinical databank driven by CGEn and COVID-19 studies from across Canada, that is broadly consented for health research. The databank contains genomes, and health metadata for 10,000+ participants affected by COVID-19. “Working on the HostSeq dataset was our plan from the start,” he says. This collaboration enabled the team to leverage a larger dataset, essential for robust conclusions.
Their findings were confirmatory – certain HLA alleles, such as C*04:01 and A*11:01, emerged as potential markers for severe COVID-19 cases (measured as the requirement for mechanical ventilation). However, the authors remain cautious, merely having these genes does not necessarily mean one will develop severe COVID-19, just that the probability is higher
What might the clinical implications of this study be? Characterizing the HLA alleles of COVID-19 patients could serve as a valuable prognostic tool. While COVID-19 is no longer a pandemic, these findings could help in guiding public health decisions for COVID-19 — such as vaccine dissemination strategies and disease management.
Using existing publicly available datasets, Warren and the research team were able to extend their expertise to address a topical and timely challenge. Looking ahead, the researchers hope to validate their findings on an even larger scale and with diverse datasets.
Footnote:
- The research story captured here was published in Establishing association between HLA-C*04:01 and severe COVID-19, HLA. 2024 Jan;103(1)
- The HostSeq databank (genomic data and health metadata) is accessible to the Canadian and international research community. The data has been consented for use in any approved health research project. More details about the phenotype variables can be found on the data portal. Currently, over 30 research projects have been approved to use this data.If you would like to learn more about HostSeq and the data access process please click here.